In genomics, big data really is big. Genomic datasets quickly consume terabytes of computer storage with more information than we have the capacity to fully analyze or understand. As a result, scientists will often publish an article about only a small piece or particular aspect of their data, leaving the rest ripe for interpretation by others. In the words, “Information isn’t the bottleneck for discovery, interpretation is.”
Although this precious genomic data is uploaded into public repositories, sadly, few people dare to touch the data because it is too complicated to understand. Data files are often mislabeled, data may be raw or analyzed and analysis from dataset to dataset is highly variable, experimental subtleties are not mentioned, and software code used to filter through data is not available.
The lack of reproducibility has far-reaching consequences.
In a recent Nature news article, Dr. Francis Collins, Director of the National Institutes of Health (NIH), and Lawrence Tabak, Deputy Director of the NIH, mention that there are “a troubling frequency of published reports that claim a significant result, but fail to be reproducible.”
Dr. John Greally, Professor at Albert Einstein College of Medicine in New York and Editor of the Genome Analysis and Tools Section of the journal Genomics, gave one example from his work. “It is impossible to truly review genomic research grant proposals,” he said, because there are so many variables in how genomic datasets are acquired, presented and analyzed. While research articles are an essential step forward in advancing science, they seem to fall short in being able to describe data well enough to make it reproducible.
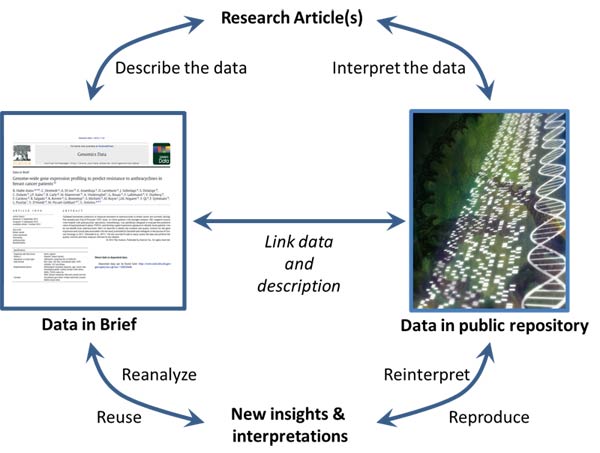
That’s where Genomics Data, one of Elsevier’s new open access journals, comes in. It provides an avenue for researchers to bring their data – along with the details necessary to understand and reuse the data – to the forefront.
The journal’s signature “Data in Brief” articles describe publicly available genomic datasets thoroughly so the data can be easily found, reproduced, reused and reanalyzed. Data in Brief are intended to supplement a research article, describing all of the nitty-gritty details that are essential to understanding the data.
Now, these kinds of accompanying details must be documented in a Specifications table at the top of each Data in Brief. The journal’s Editorial Board also checks that any related software or programming code is submitted alongside the Data in Brief.
Because Data in Brief are reviewed by the editorial board, authors receive a decision quickly: the average time for a decision on the manuscript is one week. As a result, researchers can get the word out about their datasets quickly, driving more traffic to their data and to any research article that may discuss interpretations of that data.
We hope Data in Brief will be a big step forward in making genomic data sharing and reproduction a reality.
Data in Brief — how it works
Two essential components of genomic research are:
- The data, available in a public repository: supports your research article but is not published or copyrighted as a part of that research article.
- The Research Article: an interpretation of the data.
The Data in Brief articles support these elements by providing a thorough description of the data, including quality-control checks and base-level analysis.
Data in Brief articles:
- Thoroughly describe data, facilitating reproducibility.
- Make deposited genomic data easier to find.
- Increase traffic towards associated research articles and data, leading to more citations.
- Open up doors for new collaborations.






Leave a Reply